[Paper reading] LightGBM: A Highly Efficient Gradient Boosting Decision Tree.
About three years ago you can see many top solutions of kaggle competitions used XGBoost - just one year later Microsoft proposed LightGBM - another GBM framework. And now if you go to kaggle, you can see LightGBM is gradually “replacing” XGBoost - both in top solutions and kernels. I’ve participated in about 6 competitions which require heavy feature engineerings + GBM, and I actually use XGBoost results in final submission for only once. Since LightGBM could always get same or even better performance than XGBoost in my experiences, and it runs much faster (single machine, CPU only). So this time I’ll go through its paper to see what’s special.
1. Introduction
As what we see in XGBoost paper, GBM is a powerful algorithm, but how to scale it to large and high dimensional data is a problem. Compared with vanilla GBM, XGBoost applies these techniques:
- better regularized loss function
- reasonable approximation for loss function
- shrinkage and subsampling (both sample and features)
- optimized split finding algorithms
- better lower-level system design
Well, in LightGBM paper, the authors think XGBoost’s efficiency and scalability are still unsatisfactory when handling large and high dimensional data (LOL). And they proposed more optimized approaches in split finding: Gradient based One-Side Sampling (GOSS) and Exclusive Feature Bundling (EFB). They called this new framework LightGBM. And experiments show that LightGBM speeds up the training process of conventional GBM by up to over 20 times while achieving almost the same accuracy. More detailed experiments show that LightGBM can significantly outperform XGBoost in terms of computational speed and memory consumptions.
2. Two Important Techniques
The direction for optimization here are reducing sample size and feature dimensions, they could be achieved by subsampling - like Random Forest and XGBoost, they are also supported by LightGBM. Besides that, there are two novel techniques are used.
2.1 Gradient based One-Side Sampling (GOSS)
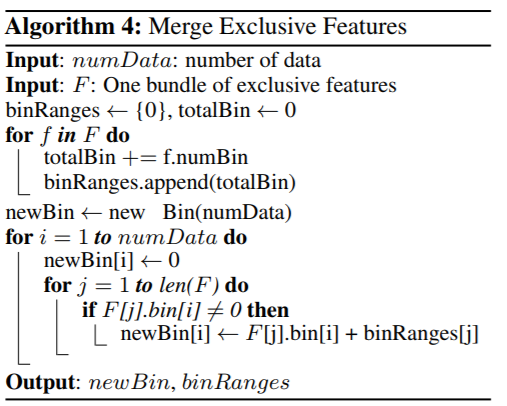
The authors notice that samples with different gradients plays different roles in computing information gain. In particular, those samples with larger gradients will contribute more to the information gain. Therefore the idea to optimize is discard samples with small gradients - since they are already well trained.
However, this will change the distribution of the data, which might hurt the model performance. Here comes the GOSS.
GOSS keeps all samples with large gradients and performs random sampling on samples with small gradients. In order to compensate the influence to the data distribution, when computing the information gain, GOSS introduces a constant multiplier for the samples with small gradients.
In detail, GOSS firstly sorts the samples according to the absolute value of the gradients and select top \(a\)% samples, then randomly sample \(b\)% samples from rest of the data. After this, amplify sampled data with small gradients by a constant \((1-a)/b\) when calculating information gain. This way for under-trained samples, the original data distribution will not change too much.
The paper also posted theoreticall analysis of this approach, I won’t get into that details here.
2.2 Exclusive Feature Bunding (EFB)
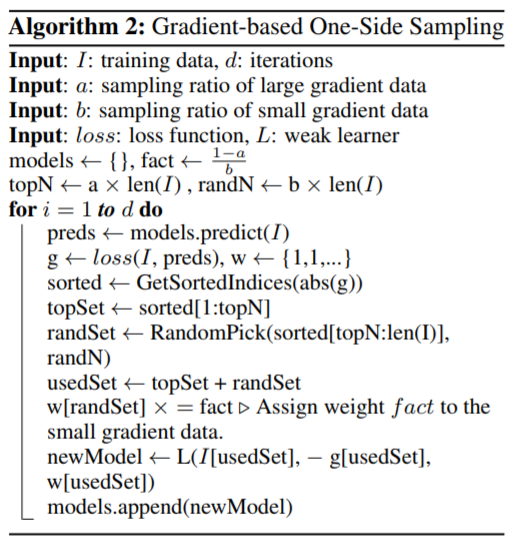
For sparse high dimensional data, it is possible to reduce the number features rather than subsampling. Specifically, in a sparse feature space, many features are mutually exlusive, which means they don’t take non-zero values at the same time (row). Then it is possible to bundle those features into a single feature. The key for this approach is to ensure that the values of original features can be identified from the bundled feature.
A simple example is: there are two features in a feature bundle, originally feature 1 takes values from \([0, 10)\) and feature 2 takes values from \([0, 20)\). Then an offset of 10 could be added to feature 2 then it will take values from \([10, 30)\). After this it is safe to merge these two features. Detailed algorithm is as this: